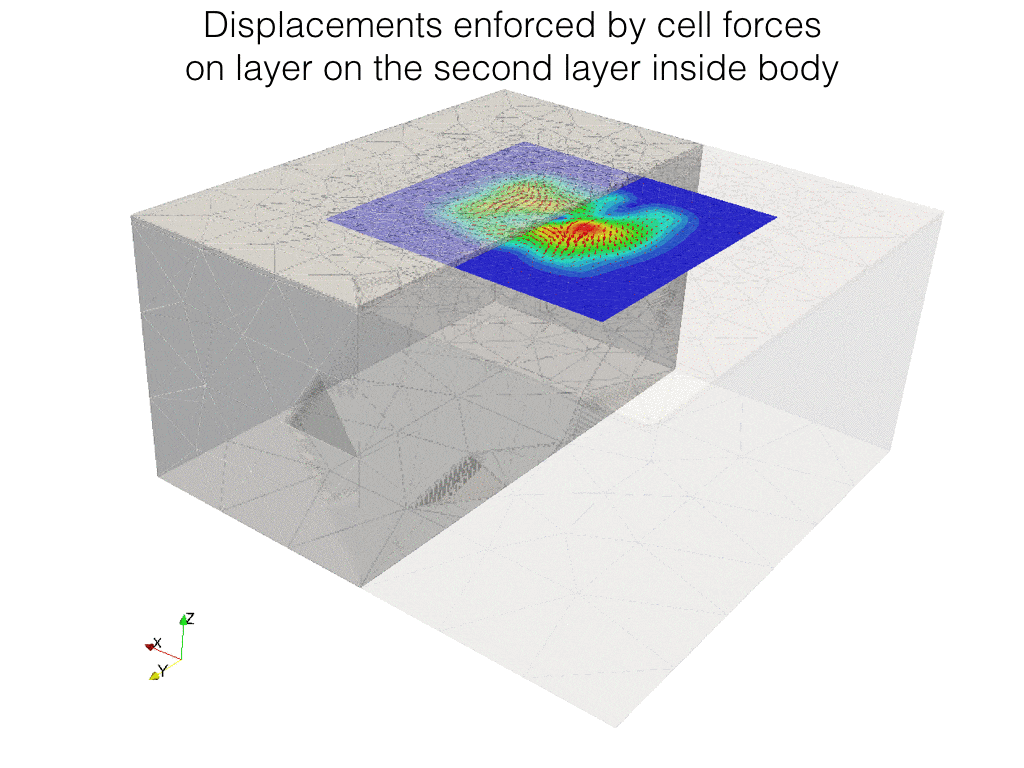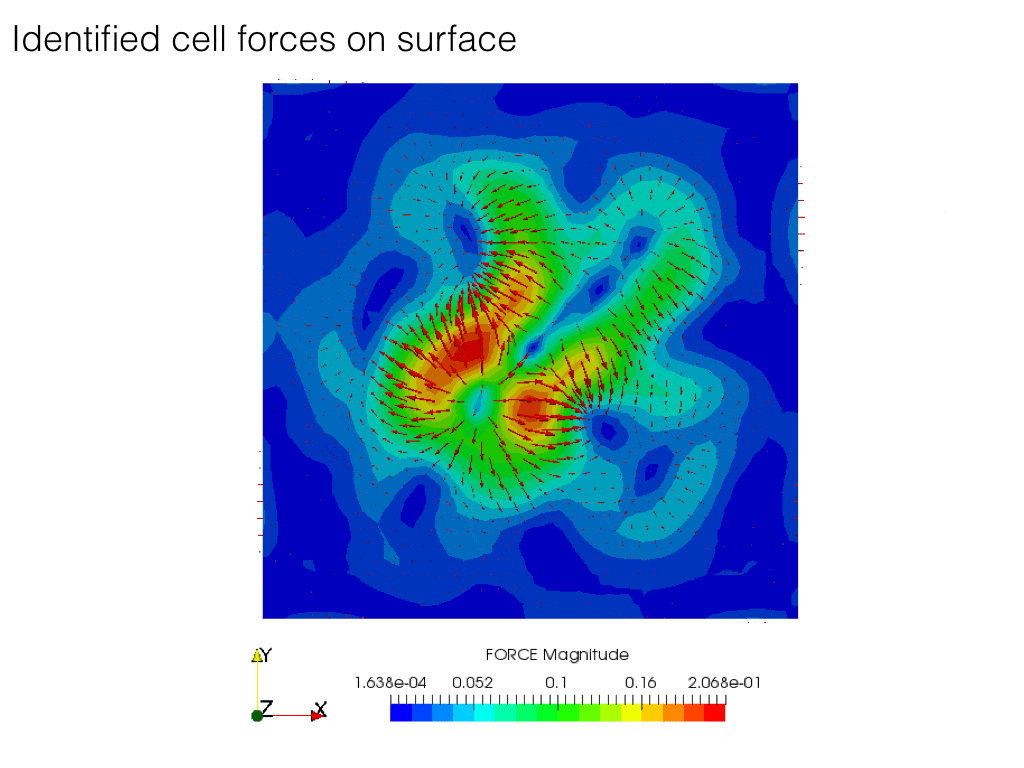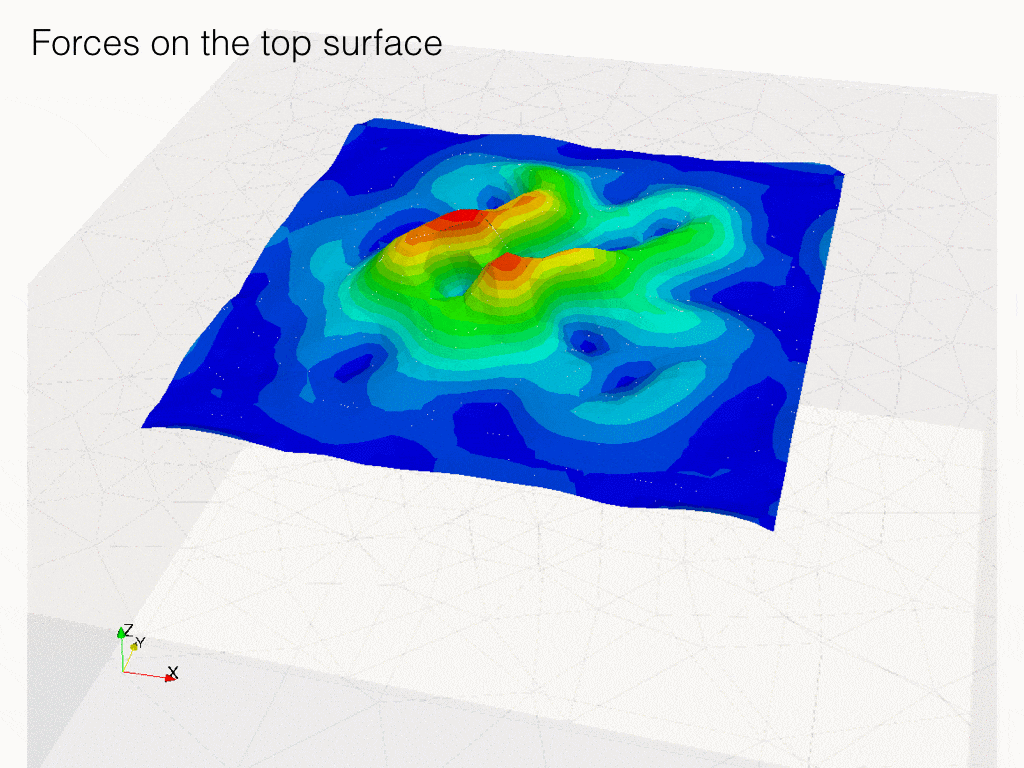
In the following code, we solve the problem of identification of forces which result in given displacements on some surface inside the body.
In the experiment, a body is covered on top by transparent gel layer, on the interface between movements of markers (bids) is observed. Strain in the body is generated by cells living on the top layer. Here we looking for forces generated by those cells.
Following implementing when needed could be easily extended to curved surfaces arbitrary located in the body.
This problem can be mathematically described by minimisation problem with the constraints, as follows
\[ \left\{ \begin{array}{l} \textrm{min}\,J(u,\rho) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} + \frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} \\ \textrm{div}[\sigma(u)] = 0 \quad \textrm{in}\,V \\ n\cdot\sigma(u) = \rho \quad \textrm{on}\,S_\rho \\ \end{array} \right. \]
Applying standard procedure, i.e. Lagrange multiplier method, we get
\[ \left\{ \begin{array}{l} \textrm{min}\,J(u,\rho,\Upsilon) = \frac{1}{2} (u,S(u-u_d))_{S_u} + \frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} + (\Upsilon,\textrm{div}[\sigma])_V \\ n\cdot\sigma = \rho \quad \textrm{on}\,S_\rho \end{array} \right. \]
\[ J(u,\rho,\Upsilon) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} +\frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} +(\textrm{grad}[\Upsilon],\sigma)_V - (\Upsilon,n\cdot\sigma)_{S_\rho} \]
and using second constraint, i.e. static constrain
\[ J(u,\rho,\Upsilon) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} +\frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} +(\textrm{grad}[\Upsilon],\sigma)_V - (\Upsilon,\rho)_{S_\rho}. \]
Note that force is enforced in week sense.
\[ (\textrm{grad}[\Upsilon],\sigma)_V = (\textrm{grad}[\Upsilon],\mathcal{A} \textrm{grad}[u] )_V = (\mathcal{A}^{*}\textrm{grad}[\Upsilon],\textrm{grad}[u] )_V = (\Sigma,\textrm{grad}[u] )_V \]
\[ J(u,\rho,\Upsilon) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} +\frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} +(\Sigma,\textrm{grad}[u])_V - (\Upsilon,\rho)_{S_\rho} \]
\[ \left\{ \begin{array}{ll} (Su,\delta u)_{S_u} + (\Sigma,\textrm{grad}[\delta u])_V &= 0 \\ (\sigma,\textrm{grad}[\delta \Upsilon])_V-(\rho,\delta \Upsilon)_{S_\rho} &= 0 \\ (\epsilon_\rho \rho,\delta \rho)_{S_\rho}-(\Upsilon,\delta \rho)_{S_\rho} &= 0 \end{array} \right. \]
and applying finite element discretization, above equations are expressed by linear algebraic equations suitable for computer calculations
\[ \left[ \begin{array}{ccc} S & K_{u \Upsilon} & 0 \\ K_{\Upsilon u} & 0 & -B^\textrm{T} \\ 0 & -B & D \end{array} \right] \left[ \begin{array}{c} u \\ \Upsilon \\ \rho \end{array} \right] = \left[ \begin{array}{c} S u_d \\ 0 \\ 0 \end{array} \right] \]
\[ \left[ \begin{array}{ccc} K & 0 & -B^\textrm{T} \\ S & K & 0 \\ 0 & -B & D \end{array} \right] \left[ \begin{array}{c} u \\ \Upsilon \\ \rho \end{array} \right] = \left[ \begin{array}{c} 0 \\ S u_d \\ 0 \end{array} \right] \]
\[ S=\epsilon_u^{-1} I\\ \epsilon_\rho^\textrm{abs} = \epsilon_\rho^\text{ref}\epsilon_u^{-1} \]
Displacement field is \(\mathbf{u}\) and \(\Upsilon\), force field on top surface is \(\boldsymbol\rho\).
We not yet explored nature of cell forces. Since the cells are attached to the body surface and anything else, the body as results those forces can not be subjected to rigid body motion.
We assume that forces are generated by 2d objects, as results only tangential forces can be produced by those forces if the surface is planar. For non-planar surfaces addition force normal to the surface is present, although the magnitude of this force could be calculated purely from geometrical considerations, thus additional physical equations are not needed.
Assuming that straight and very short pre-stressed fibres generate cell forces; a force field is curl-free. Utilising that forces can be expressed by potential field as follows
where \(\Phi\) is force potential field.
For this problem \(H^(\Omega)\) function space is required for \(u\), \(\Upsilon\) and \(\rho\). Note that \(\rho\) is given by derivative of potential field \(\Phi\) which derivatives gives potential forces.
In this variant generalisation of the local model to a weakly enforced force curl-free cell force field is considered. This generalisation is a consequence of the observation that cell has some small but finite size. Moreover, a cell has a complex pre-stressed structure which can transfer shear forces.
\[ \left\{ \begin{array}{l} \textrm{min}\,J(u,\rho) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} + \frac{\epsilon_\rho}{2}(\rho,\rho-l^2\textrm{curl}^s [\textrm{curl}^s[\rho]])_{S_\rho} \\ \textrm{div}[\sigma(u)] = 0 \quad \textrm{in}\,V \\ n\cdot\sigma = \rho \end{array} \right. \]
\[ \left\{ \begin{array}{l} \textrm{min}\,J(u,\rho) = \frac{1}{2} (u-u_d,S(u-u_d))_{S_u} + \frac{\epsilon_\rho}{2}(\rho,\rho)_{S_\rho} +\frac{\epsilon_l^{-1}}{2}(\textrm{curl}^s[\rho],\textrm{curl}^s[\rho])_{S_\rho} \\ \textrm{div}[\sigma(u)] = 0 \quad \textrm{in}\,V \\ n\cdot\sigma = \rho \end{array} \right. \]
where parameter \(\epsilon_l\) controls curl-free term, if this parameter approach zero, this formulation converge to local variant presented above.
\[ \left\{ \begin{array}{l} (Su,\delta u)_{S_u} + (\Sigma,\textrm{grad}[\delta u])_V= 0 \\ (\sigma,\textrm{grad}[\delta \Upsilon])_V-(\rho,\delta \Upsilon)_{S_\rho} = 0 \\ (\epsilon_\rho \rho,\delta \rho)_{S_\rho}+\epsilon_l^{-1}(\textrm{curl}^s[\rho],\textrm{curl}^s[\delta \rho])_{S_\rho}-(\Upsilon,\delta \rho)_{S_\rho} = 0 \end{array} \right. \]
\[ \left[ \begin{array}{ccc} S & K_{u \Upsilon} & 0 \\ K_{\Upsilon u} & 0 & -B^\textrm{T} \\ 0 & -B & D \end{array} \right] \left[ \begin{array}{c} u \\ \Upsilon \\ \rho \end{array} \right] = \left[ \begin{array}{c} S u_d \\ 0 \\ 0 \end{array} \right] \]
\[ \left[ \begin{array}{ccc} K & 0 & -B^\textrm{T} \\ S & K & 0 \\ 0 & -B & D \end{array} \right] \left[ \begin{array}{c} u \\ \Upsilon \\ \rho \end{array} \right] = \left[ \begin{array}{c} 0 \\ S u_d \\ 0 \end{array} \right] \]
For this problem \(H^(\Omega)\) function space is required for \(u\), \(\Upsilon\). Note that force field need to be in \(H(\textrm{curl},S_\rho)\) space.
If only one layer is specified, another thin polymer layer could be created automatically using prism elements,
static char help[] =
"-my_block_config set block data\n"
"\n";
int oRder;
double yOung;
double pOisson;
};
}
using namespace boost::numeric;
#include <boost/program_options.hpp>
namespace po = boost::program_options;
int main(
int argc,
char *argv[]) {
try {
PetscBool flg_block_config, flg_file;
char block_config_file[255];
PetscBool flg_order_force;
PetscInt order_force = 2;
PetscBool flg_eps_u, flg_eps_rho, flg_eps_l;
double eps_u = 1e-6;
double eps_rho = 1e-3;
double eps_l = 0;
PetscBool is_curl = PETSC_TRUE;
CHKERR PetscOptionsBegin(PETSC_COMM_WORLD,
"",
"Elastic Config",
"none");
CHKERR PetscOptionsString(
"-my_file",
"mesh file name",
"",
"mesh.h5m",
CHKERR PetscOptionsInt(
"-my_order",
"default approximation order",
"",
"-my_order_force",
"default approximation order for force approximation", "", order_force,
&order_force, &flg_order_force);
CHKERR PetscOptionsString(
"-my_block_config",
"elastic configure file name",
"", "block_conf.in", block_config_file, 255,
&flg_block_config);
CHKERR PetscOptionsReal(
"-my_eps_rho",
"foce optimality parameter ",
"",
eps_rho, &eps_rho, &flg_eps_rho);
CHKERR PetscOptionsReal(
"-my_eps_u",
"displacement optimality parameter ",
"", eps_u, &eps_u, &flg_eps_u);
CHKERR PetscOptionsReal(
"-my_eps_l",
"displacement optimality parameter ",
"", eps_l, &eps_l, &flg_eps_l);
CHKERR PetscOptionsBool(
"-my_curl",
"use Hcurl space to approximate forces",
"", is_curl, &is_curl, NULL);
ierr = PetscOptionsEnd();
if (flg_file != PETSC_TRUE) {
"*** ERROR -my_file (MESH FILE NEEDED)");
}
ParallelComm *pcomm = ParallelComm::get_pcomm(&moab,
MYPCOMM_INDEX);
if (pcomm == NULL)
pcomm = new ParallelComm(&moab, PETSC_COMM_WORLD);
const char *option;
option = "PARALLEL=READ_PART;"
"PARALLEL_RESOLVE_SHARED_ENTS;"
"PARTITION=PARALLEL_PARTITION;";
bit_level0.set(0);
{
CHKERR moab.get_entities_by_dimension(0, 3, ents3d);
}
std::vector<Range> setOrderToEnts(10);
std::map<int, BlockOptionData> block_data;
if (flg_block_config) {
double read_eps_u, read_eps_rho, read_eps_l;
try {
ifstream ini_file(block_config_file);
if (!ini_file.is_open()) {
SETERRQ(PETSC_COMM_SELF, 1,
"*** -my_block_config does not exist ***");
}
po::variables_map vm;
po::options_description config_file_options;
config_file_options.add_options()(
"eps_u", po::value<double>(&read_eps_u)->default_value(-1))(
"eps_rho", po::value<double>(&read_eps_rho)->default_value(-1))(
"eps_l", po::value<double>(&read_eps_l)->default_value(-1));
std::ostringstream str_order;
str_order << "block_" << it->getMeshsetId() << ".displacement_order";
config_file_options.add_options()(
str_order.str().c_str(),
po::value<int>(&block_data[it->getMeshsetId()].oRder)
std::ostringstream str_cond;
str_cond << "block_" << it->getMeshsetId() << ".young_modulus";
config_file_options.add_options()(
str_cond.str().c_str(),
po::value<double>(&block_data[it->getMeshsetId()].yOung)
->default_value(-1));
std::ostringstream str_capa;
str_capa << "block_" << it->getMeshsetId() << ".poisson_ratio";
config_file_options.add_options()(
str_capa.str().c_str(),
po::value<double>(&block_data[it->getMeshsetId()].pOisson)
->default_value(-2));
}
po::parsed_options parsed =
parse_config_file(ini_file, config_file_options, true);
store(parsed, vm);
po::notify(vm);
if (block_data[it->getMeshsetId()].oRder == -1)
continue;
if (block_data[it->getMeshsetId()].oRder ==
order)
continue;
PetscPrintf(PETSC_COMM_WORLD, "Set block %d order to %d\n",
it->getMeshsetId(), block_data[it->getMeshsetId()].oRder);
CHKERR moab.get_entities_by_handle(it->getMeshset(), block_ents,
true);
CHKERR moab.get_connectivity(block_ents, nodes,
true);
Range ents_to_set_order, ents3d;
CHKERR moab.get_adjacencies(nodes, 3,
false, ents3d,
moab::Interface::UNION);
CHKERR moab.get_adjacencies(ents3d, 2,
false, ents_to_set_order,
moab::Interface::UNION);
CHKERR moab.get_adjacencies(ents3d, 1,
false, ents_to_set_order,
moab::Interface::UNION);
ents_to_set_order = subtract(
ents_to_set_order, ents_to_set_order.subset_by_type(MBQUAD));
ents_to_set_order = subtract(
ents_to_set_order, ents_to_set_order.subset_by_type(MBPRISM));
set_order_ents.merge(ents3d);
set_order_ents.merge(ents_to_set_order);
setOrderToEnts[block_data[it->getMeshsetId()].oRder].merge(
set_order_ents);
}
std::vector<std::string> additional_parameters;
additional_parameters =
collect_unrecognized(parsed.options, po::include_positional);
for (std::vector<std::string>::iterator vit =
additional_parameters.begin();
vit != additional_parameters.end(); vit++) {
CHKERR PetscPrintf(PETSC_COMM_WORLD,
"** WARNING Unrecognized option %s\n",
vit->c_str());
}
} catch (const std::exception &ex) {
std::ostringstream ss;
ss << ex.what() << std::endl;
}
if (read_eps_u > 0) {
eps_u = read_eps_u;
};
if (read_eps_rho > 0) {
eps_rho = read_eps_rho;
}
if (read_eps_l > 0) {
eps_l = read_eps_l;
}
}
PetscPrintf(PETSC_COMM_WORLD, "epsU = %6.4e epsRho = %6.4e\n", eps_u,
eps_rho);
if (is_curl) {
} else {
}
ents_1st_layer, true);
vertex_to_fix, false);
false);
if (vertex_to_fix.size() != 1 && !vertex_to_fix.empty()) {
"Should be one vertex only, but is %d", vertex_to_fix.size());
}
}
ents_1st_layer.subset_by_type(MBTRI), MBTRI, "RHO");
ents_2nd_layer, true);
}
for (int oo = 2; oo != setOrderToEnts.size(); oo++) {
if (setOrderToEnts[oo].size() > 0) {
}
}
const int through_thickness_order = 2;
{
CHKERR moab.get_entities_by_dimension(0, 3, ents3d);
CHKERR moab.get_adjacencies(ents3d, 2,
false, ents,
moab::Interface::UNION);
CHKERR moab.get_adjacencies(ents3d, 1,
false, ents,
moab::Interface::UNION);
CHKERR moab.get_entities_by_type(0, MBPRISM, prisms);
{
CHKERR moab.get_adjacencies(prisms, 2,
false, quads,
moab::Interface::UNION);
prism_tris = quads.subset_by_type(MBTRI);
quads = subtract(quads, prism_tris);
CHKERR moab.get_adjacencies(quads, 1,
false, quads_edges,
moab::Interface::UNION);
CHKERR moab.get_adjacencies(prism_tris, 1,
false, prism_tris_edges,
moab::Interface::UNION);
quads_edges = subtract(quads_edges, prism_tris_edges);
prisms.merge(quads);
prisms.merge(quads_edges);
}
ents.merge(ents3d);
ents = subtract(ents, set_order_ents);
ents = subtract(ents, prisms);
through_thickness_order);
}
if (is_curl) {
} else {
}
boost::shared_ptr<Hooke<adouble>> hooke_adouble_ptr(
new Hooke<adouble>());
boost::shared_ptr<Hooke<double>> hooke_double_ptr(
new Hooke<double>());
CHKERR elastic.setBlocks(hooke_double_ptr, hooke_adouble_ptr);
CHKERR elastic.addElement(
"ELASTIC",
"U");
CHKERR elastic.setOperators(
"U",
"MESH_NODE_POSITIONS",
false,
true);
{
elastic.setOfBlocks[2].tEts);
elastic.setOfBlocks[2].tEts, MBPRISM, "ELASTIC_PRISM");
fat_prism_rhs.getOpPtrVector().push_back(
fat_prism_rhs.getOpPtrVector().push_back(
fat_prism_rhs.getOpPtrVector().push_back(
"U", elastic.commonData));
fat_prism_rhs.getOpPtrVector().push_back(
"U", elastic.setOfBlocks[2], elastic.commonData, 2, false, false,
true));
fat_prism_rhs.getOpPtrVector().push_back(
"U", elastic.setOfBlocks[2], elastic.commonData));
fat_prism_lhs.getOpPtrVector().push_back(
fat_prism_lhs.getOpPtrVector().push_back(
fat_prism_lhs.getOpPtrVector().push_back(
"U", elastic.commonData));
fat_prism_lhs.getOpPtrVector().push_back(
"U", elastic.setOfBlocks[2], elastic.commonData, 2, true, false,
true));
fat_prism_lhs.getOpPtrVector().push_back(
"U", "U", elastic.setOfBlocks[2], elastic.commonData));
}
int id = it->getMeshsetId();
if (block_data[id].yOung > 0) {
elastic.setOfBlocks[id].E = block_data[id].yOung;
CHKERR PetscPrintf(PETSC_COMM_WORLD,
"Block %d set Young modulus %3.4g\n", id,
elastic.setOfBlocks[id].E);
}
if (block_data[id].pOisson >= -1) {
elastic.setOfBlocks[id].PoissonRatio = block_data[id].pOisson;
CHKERR PetscPrintf(PETSC_COMM_WORLD,
"Block %d set Poisson ratio %3.4g\n", id,
elastic.setOfBlocks[id].PoissonRatio);
}
}
"UPSILON");
"U");
"UPSILON");
"U");
"DISP_X");
"DISP_Y");
"DISPLACEMENTS_PENALTY");
"BT");
"B");
"D");
DMType dm_name = "MOFEM";
DM dm_control;
{
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_control);
CHKERR DMSetType(dm_control, dm_name);
bit_level0);
CHKERR DMSetFromOptions(dm_control);
}
ublas::matrix<Mat> nested_matrices(2, 2);
ublas::vector<IS> nested_is_rows(2);
ublas::vector<IS> nested_is_cols(2);
for (
int i = 0;
i != 2;
i++) {
nested_is_rows[
i] = PETSC_NULL;
nested_is_cols[
i] = PETSC_NULL;
for (
int j = 0;
j != 2;
j++) {
nested_matrices(
i,
j) = PETSC_NULL;
}
}
ublas::matrix<Mat> sub_nested_matrices(2, 2);
ublas::vector<IS> sub_nested_is_rows(2);
ublas::vector<IS> sub_nested_is_cols(2);
for (
int i = 0;
i != 2;
i++) {
sub_nested_is_rows[
i] = PETSC_NULL;
sub_nested_is_cols[
i] = PETSC_NULL;
for (
int j = 0;
j != 2;
j++) {
sub_nested_matrices(
i,
j) = PETSC_NULL;
}
}
DM dm_sub_volume_control;
{
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_sub_volume_control);
CHKERR DMSetType(dm_sub_volume_control, dm_name);
"SUB_CONTROL_PROB");
CHKERR DMSetUp(dm_sub_volume_control);
boost::shared_ptr<Problem::SubProblemData> sub_data =
CHKERR sub_data->getRowIs(&nested_is_rows[0]);
CHKERR sub_data->getColIs(&nested_is_cols[0]);
nested_matrices(0, 0) = PETSC_NULL;
}
{
DM dm_sub_sub_elastic;
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_sub_sub_elastic);
CHKERR DMSetType(dm_sub_sub_elastic, dm_name);
"ELASTIC_PROB");
CHKERR DMSetUp(dm_sub_sub_elastic);
Mat Kuu;
CHKERR DMCreateMatrix(dm_sub_sub_elastic, &Kuu);
CHKERR DMCreateGlobalVector(dm_sub_sub_elastic, &Du);
CHKERR DMCreateGlobalVector(dm_sub_sub_elastic, &Fu);
SCATTER_REVERSE);
boost::shared_ptr<FEMethod> dirihlet_bc_ptr;
dirihlet_bc_ptr = boost::shared_ptr<FEMethod>(
dirihlet_bc_ptr.get());
elastic.getLoopFeRhs().snes_f = Fu;
fat_prism_rhs.snes_f = Fu;
&elastic.getLoopFeRhs());
&fat_prism_rhs);
elastic.getLoopFeLhs().snes_B = Kuu;
fat_prism_lhs.snes_B = Kuu;
&elastic.getLoopFeLhs());
&fat_prism_lhs);
dirihlet_bc_ptr.get());
CHKERR VecGhostUpdateBegin(Fu, ADD_VALUES, SCATTER_REVERSE);
CHKERR VecGhostUpdateEnd(Fu, ADD_VALUES, SCATTER_REVERSE);
boost::shared_ptr<Problem::SubProblemData> sub_data =
CHKERR sub_data->getRowIs(&sub_nested_is_rows[0]);
CHKERR sub_data->getColIs(&sub_nested_is_cols[0]);
sub_nested_matrices(0, 0) = Kuu;
IS isUpsilon;
->isCreateFromProblemFieldToOtherProblemField(
"ELASTIC_PROB",
"U",
ROW,
"SUB_CONTROL_PROB",
"UPSILON",
ROW,
PETSC_NULL, &isUpsilon);
sub_nested_is_rows[1] = isUpsilon;
sub_nested_is_cols[1] = isUpsilon;
sub_nested_matrices(1, 1) = Kuu;
PetscObjectReference((PetscObject)Kuu);
PetscObjectReference((PetscObject)isUpsilon);
cerr << "Kuu" << endl;
MatView(Kuu, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
CHKERR DMDestroy(&dm_sub_sub_elastic);
}
{
DM dm_sub_disp_penalty;
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_sub_disp_penalty);
CHKERR DMSetType(dm_sub_disp_penalty, dm_name);
"S_PROB");
CHKERR DMSetUp(dm_sub_disp_penalty);
Mat S;
CHKERR DMCreateMatrix(dm_sub_disp_penalty, &S);
face_element.getOpPtrVector().push_back(
new OpCellS(S, eps_u));
"DISPLACEMENTS_PENALTY", &face_element);
CHKERR MatAssemblyBegin(S, MAT_FLUSH_ASSEMBLY);
CHKERR MatAssemblyEnd(S, MAT_FLUSH_ASSEMBLY);
cerr << "S" << endl;
MatView(S, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
boost::shared_ptr<Problem::SubProblemData> sub_data =
sub_nested_matrices(1, 0) = S;
CHKERR DMDestroy(&dm_sub_disp_penalty);
}
{
DM dm_sub_force_penalty;
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_sub_force_penalty);
CHKERR DMSetType(dm_sub_force_penalty, dm_name);
CHKERR DMSetUp(dm_sub_force_penalty);
CHKERR DMCreateMatrix(dm_sub_force_penalty, &
D);
{
face_d_matrix.getOpPtrVector().push_back(
new OpCalculateInvJacForFace(inv_jac));
if (is_curl) {
face_d_matrix.getOpPtrVector().push_back(
face_d_matrix.getOpPtrVector().push_back(
} else {
face_d_matrix.getOpPtrVector().push_back(
face_d_matrix.getOpPtrVector().push_back(
}
&face_d_matrix);
}
CHKERR MatAssemblyBegin(
D, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyEnd(
D, MAT_FINAL_ASSEMBLY);
if (is_curl == PETSC_FALSE) {
int nb_dofs_to_fix = 0;
int index_to_fix = 0;
if (!vertex_to_fix.empty()) {
boost::shared_ptr<NumeredDofEntity> dof_ptr;
dof_ptr);
if (dof_ptr) {
nb_dofs_to_fix = 1;
index_to_fix = dof_ptr->getPetscGlobalDofIdx();
cerr << *dof_ptr << endl;
}
}
}
CHKERR MatZeroRowsColumns(
D, nb_dofs_to_fix, &index_to_fix,
eps_rho / eps_u, PETSC_NULL, PETSC_NULL);
} else {
std::vector<int> dofs_to_fix;
for (auto p_eit = edges_to_fix.pair_begin();
p_eit != edges_to_fix.pair_end(); ++p_eit) {
auto lo = row_dofs->lower_bound(
auto hi =
bit_number, p_eit->second));
for (; lo != hi; ++lo)
dofs_to_fix.push_back((*lo)->getPetscGlobalDofIdx());
}
CHKERR MatZeroRowsColumns(
D, dofs_to_fix.size(), &*dofs_to_fix.begin(),
eps_rho / eps_u, PETSC_NULL, PETSC_NULL);
}
cerr << "D" << endl;
MatView(
D, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
boost::shared_ptr<Problem::SubProblemData> sub_data =
CHKERR sub_data->getRowIs(&nested_is_rows[1]);
CHKERR sub_data->getColIs(&nested_is_cols[1]);
nested_matrices(1, 1) =
D;
CHKERR DMDestroy(&dm_sub_force_penalty);
}
{
DM dm_sub_force;
CHKERR DMCreate(PETSC_COMM_WORLD, &dm_sub_force);
CHKERR DMSetType(dm_sub_force, dm_name);
Mat UB, UPSILONB;
CHKERR DMCreateMatrix(dm_sub_force, &UB);
CHKERR DMCreateMatrix(dm_sub_force, &UPSILONB);
CHKERR MatZeroEntries(UPSILONB);
{
face_b_matrices.getOpPtrVector().push_back(
new OpCalculateInvJacForFace(inv_jac));
if (is_curl) {
face_b_matrices.getOpPtrVector().push_back(
face_b_matrices.getOpPtrVector().push_back(
face_b_matrices.getOpPtrVector().push_back(
new OpCellCurlB(UB,
"U"));
face_b_matrices.getOpPtrVector().push_back(
} else {
face_b_matrices.getOpPtrVector().push_back(
face_b_matrices.getOpPtrVector().push_back(
face_b_matrices.getOpPtrVector().push_back(
}
}
CHKERR MatAssemblyBegin(UB, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyBegin(UPSILONB, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyEnd(UB, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyEnd(UPSILONB, MAT_FINAL_ASSEMBLY);
if (is_curl == PETSC_FALSE) {
int nb_dofs_to_fix = 0;
int index_to_fix = 0;
if (!vertex_to_fix.empty()) {
boost::shared_ptr<NumeredDofEntity> dof_ptr;
dof_ptr);
if (dof_ptr) {
nb_dofs_to_fix = 1;
index_to_fix = dof_ptr->getPetscGlobalDofIdx();
cerr << *dof_ptr << endl;
}
}
}
CHKERR MatZeroRows(UB, nb_dofs_to_fix, &index_to_fix, 0, PETSC_NULL,
PETSC_NULL);
CHKERR MatZeroRows(UPSILONB, nb_dofs_to_fix, &index_to_fix, 0,
PETSC_NULL, PETSC_NULL);
} else {
std::vector<int> dofs_to_fix;
for (auto p_eit = edges_to_fix.pair_begin();
p_eit != edges_to_fix.pair_end(); ++p_eit) {
auto lo = row_dofs->lower_bound(
auto hi =
bit_number, p_eit->second));
for (; lo != hi; ++lo)
dofs_to_fix.push_back((*lo)->getPetscGlobalDofIdx());
}
CHKERR MatZeroRows(UB, dofs_to_fix.size(), &*dofs_to_fix.begin(), 0,
PETSC_NULL, PETSC_NULL);
CHKERR MatZeroRows(UPSILONB, dofs_to_fix.size(), &*dofs_to_fix.begin(),
0, PETSC_NULL, PETSC_NULL);
}
Mat UBT;
CHKERR MatTranspose(UB, MAT_INITIAL_MATRIX, &UBT);
cerr << "UBT" << endl;
MatView(UBT, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
boost::shared_ptr<Problem::SubProblemData> sub_data =
nested_matrices(0, 1) = UBT;
cerr << "UPSILONB" << endl;
MatView(UPSILONB, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
nested_matrices(1, 0) = UPSILONB;
CHKERR DMDestroy(&dm_sub_force);
}
Mat SubA;
CHKERR MatCreateNest(PETSC_COMM_WORLD, 2, &sub_nested_is_rows[0], 2,
&sub_nested_is_cols[0], &sub_nested_matrices(0, 0),
&SubA);
nested_matrices(0, 0) = SubA;
CHKERR MatAssemblyBegin(SubA, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyEnd(SubA, MAT_FINAL_ASSEMBLY);
cerr << "Nested SubA" << endl;
MatView(SubA, PETSC_VIEWER_STDOUT_WORLD);
}
CHKERR MatCreateNest(PETSC_COMM_WORLD, 2, &nested_is_rows[0], 2,
&nested_is_cols[0], &nested_matrices(0, 0), &
A);
CHKERR MatAssemblyBegin(
A, MAT_FINAL_ASSEMBLY);
CHKERR MatAssemblyEnd(
A, MAT_FINAL_ASSEMBLY);
cerr << "Nested A" << endl;
MatView(
A, PETSC_VIEWER_STDOUT_WORLD);
}
CHKERR DMCreateGlobalVector(dm_control, &
D);
CHKERR DMCreateGlobalVector(dm_control, &
F);
{
face_element.getOpPtrVector().push_back(
new OpGetDispX(common_data));
face_element.getOpPtrVector().push_back(
new OpGetDispY(common_data));
face_element.getOpPtrVector().push_back(
&face_element);
CHKERR VecGhostUpdateBegin(
F, ADD_VALUES, SCATTER_REVERSE);
CHKERR VecGhostUpdateEnd(
F, ADD_VALUES, SCATTER_REVERSE);
}
KSP solver;
{
CHKERR KSPCreate(PETSC_COMM_WORLD, &solver);
CHKERR KSPSetDM(solver, dm_control);
CHKERR KSPSetFromOptions(solver);
CHKERR KSPSetDMActive(solver, PETSC_FALSE);
CHKERR KSPSetInitialGuessKnoll(solver, PETSC_FALSE);
CHKERR KSPSetInitialGuessNonzero(solver, PETSC_FALSE);
PC pc;
CHKERR PCSetType(pc, PCFIELDSPLIT);
PetscBool is_pcfs = PETSC_FALSE;
PetscObjectTypeCompare((PetscObject)pc, PCFIELDSPLIT, &is_pcfs);
if (is_pcfs) {
CHKERR PCFieldSplitSetIS(pc, NULL, nested_is_rows[0]);
CHKERR PCFieldSplitSetIS(pc, NULL, nested_is_rows[1]);
CHKERR PCFieldSplitSetType(pc, PC_COMPOSITE_SCHUR);
KSP *sub_ksp;
CHKERR PCFieldSplitGetSubKSP(pc, &
n, &sub_ksp);
{
PC sub_pc_0;
CHKERR KSPGetPC(sub_ksp[0], &sub_pc_0);
CHKERR PCSetOperators(sub_pc_0, SubA, SubA);
CHKERR PCSetType(sub_pc_0, PCFIELDSPLIT);
CHKERR PCFieldSplitSetIS(sub_pc_0, NULL, sub_nested_is_rows[0]);
CHKERR PCFieldSplitSetIS(sub_pc_0, NULL, sub_nested_is_rows[1]);
CHKERR PCFieldSplitSetType(sub_pc_0, PC_COMPOSITE_MULTIPLICATIVE);
}
} else {
"Only works with pre-conditioner PCFIELDSPLIT");
}
}
CHKERR VecGhostUpdateBegin(
D, INSERT_VALUES, SCATTER_FORWARD);
CHKERR VecGhostUpdateEnd(
D, INSERT_VALUES, SCATTER_FORWARD);
SCATTER_REVERSE);
CHKERR VecView(
D, PETSC_VIEWER_DRAW_WORLD);
std::string wait;
std::cin >> wait;
}
for (
int i = 0;
i != 2;
i++) {
if (sub_nested_is_rows[
i]) {
CHKERR ISDestroy(&sub_nested_is_rows[
i]);
}
if (sub_nested_is_cols[
i]) {
CHKERR ISDestroy(&sub_nested_is_cols[
i]);
}
for (
int j = 0;
j != 2;
j++) {
if (sub_nested_matrices(
i,
j)) {
CHKERR MatDestroy(&sub_nested_matrices(
i,
j));
}
}
}
for (
int i = 0;
i != 2;
i++) {
CHKERR ISDestroy(&nested_is_rows[
i]);
}
CHKERR ISDestroy(&nested_is_cols[
i]);
}
for (
int j = 0;
j != 2;
j++) {
if (nested_matrices(
i,
j)) {
CHKERR MatDestroy(&nested_matrices(
i,
j));
}
}
}
CHKERR DMDestroy(&dm_sub_volume_control);
{
CHKERR post_proc.generateReferenceElementMesh();
CHKERR post_proc.addFieldValuesPostProc(
"U");
CHKERR post_proc.addFieldValuesGradientPostProc(
"U");
m_field, post_proc.postProcMesh, post_proc.mapGaussPts, "U",
post_proc.commonData, &elastic.setOfBlocks));
CHKERR post_proc.writeFile(
"out.h5m");
elastic.getLoopFeEnergy().eNergy = 0;
&elastic.getLoopFeEnergy());
PetscPrintf(PETSC_COMM_WORLD, "Elastic energy %6.4e\n",
elastic.getLoopFeEnergy().eNergy);
}
{
CHKERR post_proc_face.generateReferenceElementMesh();
post_proc_face.getOpPtrVector().push_back(
new OpCalculateInvJacForFace(inv_jac));
if (is_curl) {
post_proc_face.getOpPtrVector().push_back(
post_proc_face.getOpPtrVector().push_back(
} else {
post_proc_face.getOpPtrVector().push_back(
post_proc_face.getOpPtrVector().push_back(
}
post_proc_face.getOpPtrVector().push_back(
post_proc_face.mapGaussPts, common_data));
CHKERR post_proc_face.writeFile(
"out_tractions.h5m");
}
CHKERR DMDestroy(&dm_control);
}
return 0;
}